COMPUTATIONAL ANATOMY
We examined the reproducibility and the power to detect real changes of different computational techniques. It is the first work to systematically investigate the reproducibility and variability of different registration methods in tensor-based morphometry (TBM). In particular, we compared matching functionals (sum of squared differences (L2) and mutual information (MI)), as well as large deformation registration schemes (unbiased registration and viscous fluid registration) using serial MRI scans of ten normal elderly patients from the preparatory phase of the Alzheimer's Disease Neuroimaging Initiative (ADNI) and ten Alzheimer's subjects from the ADNI follow-up phase. Our results show that the unbiased methods have higher reproducibility. The unbiased methods are less likely to produce changes in the absence of any real physiological change. Moreover, they are also better in detecting biological deformations by penalizing any bias in the corresponding statistical maps.
FOLLOW-UP SCANS
 |
 |
 |
 |
||
 |
 |
 |
 |
||
| L2-FLUID | L2-UNBIASED | MI-FLUID | MI-UNBIASED | ||
 |
|||||
|
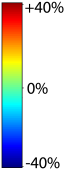
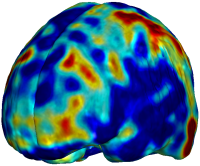
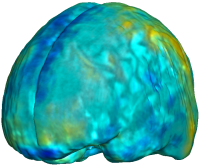
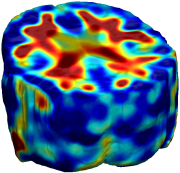
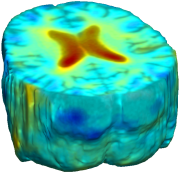
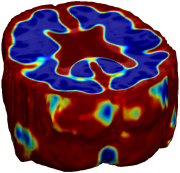
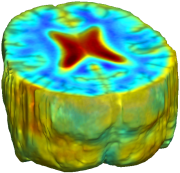
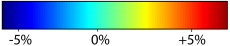
Nonrigid registration was performed on the ADNI Follow-up study (serial MRI images acquired one year apart) using L2-Fluid, L2-Unbiased, MI-Fluid, and MI-Unbiased registration methods. For each method, the mean of the resulting ten Jacobian maps is superimposed on one of the brain volumes. Visually, Fluid registration generates noisy mean maps, while maps generated using Unbiased methods suggest a volume reduction in gray matter as well as ventricle enlargement. |
|||||
BASELINE SCANS
 |
 |
 |
 |
||
 |
 |
 |
 |
||
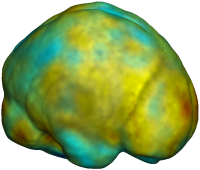
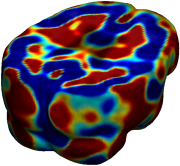
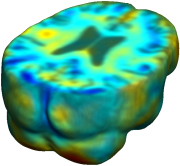
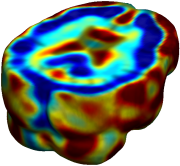
| L2-FLUID | L2-UNBIASED | MI-FLUID | MI-UNBIASED | ||
 |
|||||
|
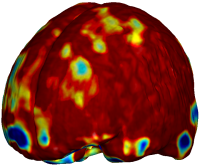
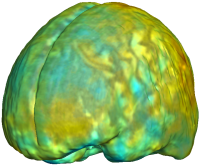
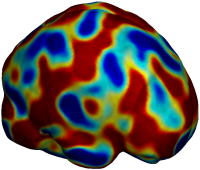
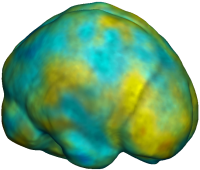
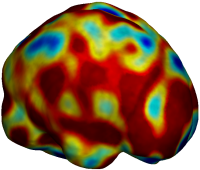
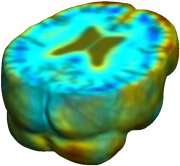
Nonrigid registration was performed on the ADNI Baseline study (serial MRI images acquired two weeks apart) of ten normal elderly subjects using L2-Fluid, L2-Unbiased, MI-Fluid, and MI-Unbiased registration methods. For each method, the mean of the resulting ten Jacobian maps is superimposed on one of the brain volumes. Visually, Fluid registration generates noisy mean maps, while maps generated using Unbiased methods are less noisy with values closer to 1. |
|||||
INVERSE CONSISTENCY OF UNBIASED REGISTRATION
| time 2 to time 1 |
 |
 |
 |
 |
|
| time 1 to time 2 |
 |
 |
 |
 |
|
| Jacobian products |
 |
 |
 |
 |
|
| L2-FLUID | L2-UNBIASED | MI-FLUID | MI-UNBIASED | ||
|
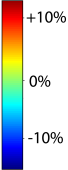
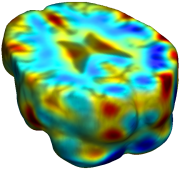
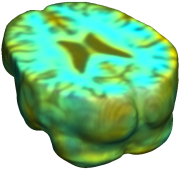
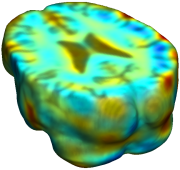
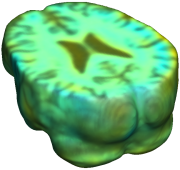
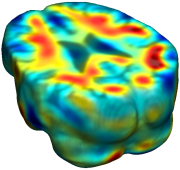
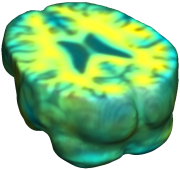
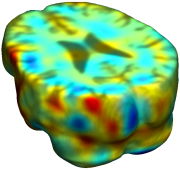
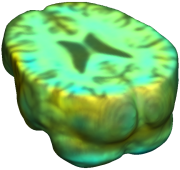
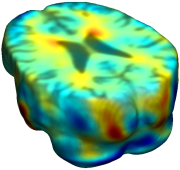
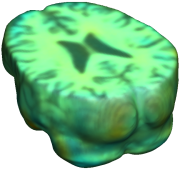
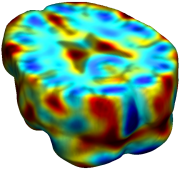
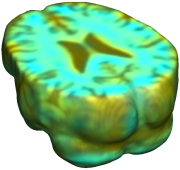
This figure examines the inverse consistency of deformation models. Nonrigid registration was performed on an image pair from one of the subjects from the ADNI Baseline study (serial MRI images acquired two weeks apart) using L2-Fluid, L2-Unbiased, MI-Fluid, and MI-Unbiased registration methods. Jacobian maps of deformations from time 2 to time 1 (row 1) and time 1 to time 2 (row 2) are superimposed on the target volumes. The unbiased methods generate less noisy Jacobian maps with values closer to 1; this shows the greater stability of the approach when no volumetric change is present. Products of Jacobian maps (row 3) are shown for the forward and backward directions. For the unbiased methods, the products of the Jacobian maps are less noisy, with values closer to 1, showing better inverse consistency. |
|||||
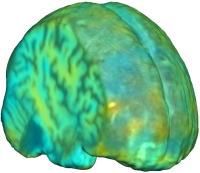
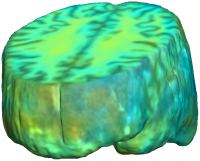
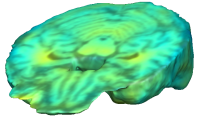
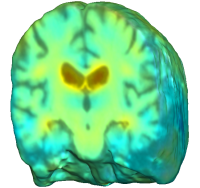
UNBIASED REGISTRATION FOR DETECTING ALZHEIMER'S DISEASE
| CTL |
 |
 |
 |
 |
|
| MCI |
 |
 |
 |
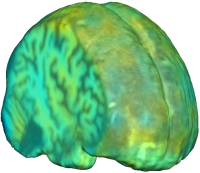 |
|
| AD |
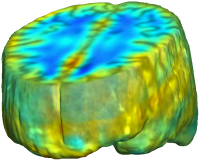 |
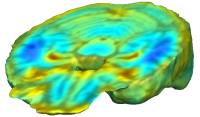 |
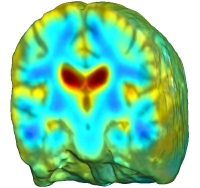 |
 |
|
 |
|||||
|
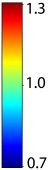
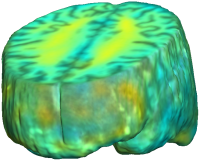
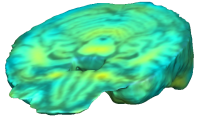
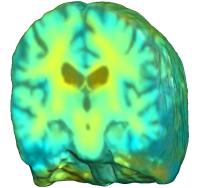
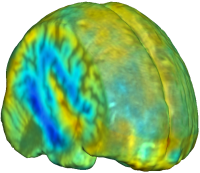
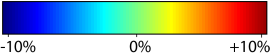
Unbiased registration with L2 matching was performed on 100 pairs of serial MR images, acquired 12 months apart, from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) dataset. The selected sample consisted of 20 patients with Alzheimer’s disease (AD), 40 individuals with mild cognitive impairment (MCI), and 40 healthy elderly controls (CTL). The mean of the resulting Jacobian maps in each group is superimposed on a brain volume. |
|||||
|
|
|||||
|
References: Igor Yanovsky, Alex Leow, Suh Lee, Stanley Osher, Paul Thompson, Comparing Registration Methods for Mapping Brain Change using Tensor-Based Morphometry, Medical Image Analysis, vol. 13, no. 5, 679-700, 2009. Igor Yanovsky, Paul Thompson, Stanley Osher, Alex Leow, Asymmetric and Symmetric Unbiased Image Registration: Statistical Assessment of Performance, Mathematical Methods in Biomedical Image Analysis, 2008. |
|||||
UNBIASED NONLINEAR ELASTIC REGISTRATION
We propose a new nonlinear image registration model which is based on nonlinear elastic regularization and unbiased registration. The unbiased large-deformation nonlinear elasticity method was tested using volumetric serial magnetic resonance images and shown to have some advantages for medical imaging applications.
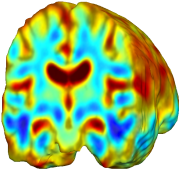
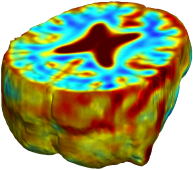
| Unbiased Fluid with L2 |
 |
 |
 |
 |
|
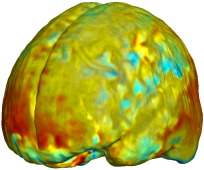
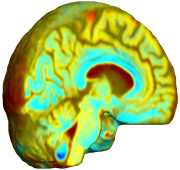
| Unbiased Nonlinear Elasticity with L2 |
 |
 |
 |
 |
|
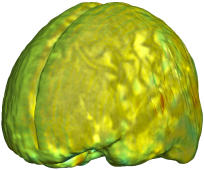
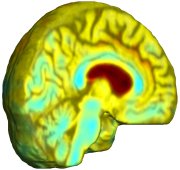
| Unbiased Fluid with MI |
 |
 |
 |
 |
|
| Unbiased Nonlinear Elasticity with MI |
 |
 |
 |
 |
|
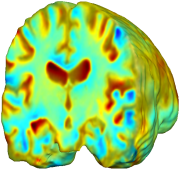
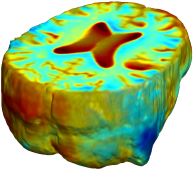
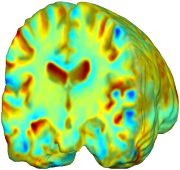
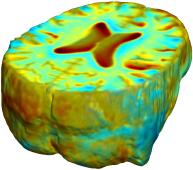
| L2-FLUID | L2-UNBIASED | MI-FLUID | MI-UNBIASED | ||
 |
|||||
|
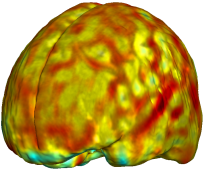
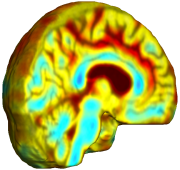
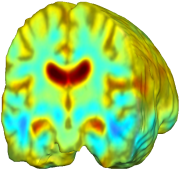
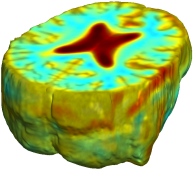
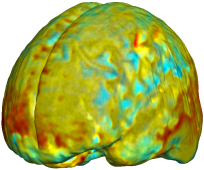
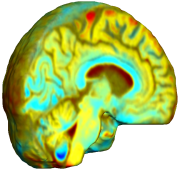
Nonrigid registration was performed on the Serial MRI images from the ADNI Follow-up dataset using unbiased fluid registration and unbiased nonlinear elasticity registration, both coupled with L2 and MI matching. Jacobian maps are superimposed on the target volume. |
|||||
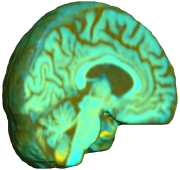
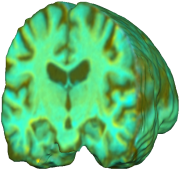
INVERSE CONSISTENCY OF UNBIASED NONLINEAR ELASTIC REGISTRATION
| time 2 to time 1 |
 |
 |
 |
 |
|
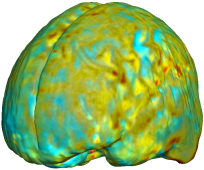
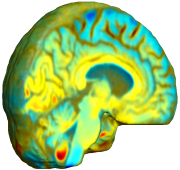
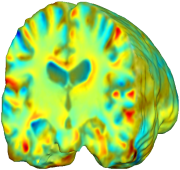
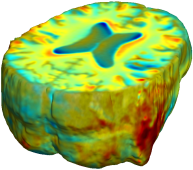
| time 1 to time 2 |
 |
 |
 |
 |
|
| Jacobian products |
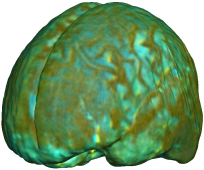 |
 |
 |
 |
|
|
This figure examines the inverse consistency of the unbiased nonlinear elastic registration. Here, the model is coupled with mutual information matching. Jacobian maps of deformations from time 2 to time 1 (row 1) and time 1 to time 2 (row 2) are superimposed on the target volumes. The products of Jacobian maps, shown in row 3, have values close to 1, suggesting inverse consistency. |
|||||
|
References: Igor Yanovsky, Carole Le Guyader, Alex Leow, Paul Thompson, Luminita Vese, Nonlinear Elastic Registration with Unbiased Regularization in Three Dimensions, The MIDAS Journal (549), 56–67, http://hdl.handle.net/10380/1360, 2008. |
|||||